A Modular, Dynamic, DNA-Based Platform for Regulating Cargo Distribution and Transport Between Lipid Domains
Nano Letters(2021)SCI 1区SCI 2区
The authors of this paper include Roger Rubio-Sánchez, affiliated with the Department of Biology and Soft Systems at the Cavendish Laboratory, University of Cambridge; Simone Eizagirre Barker, a PhD student at the Centre for Doctoral Training in Nanoscience and Nanotechnology, University of Cambridge, whose research focuses on the magneto-optical properties of novel quantum luminescent materials; Michał Walczak, employed at the Institute of Human Genetics, Polish Academy of Sciences; Pietro Cicuta, from the Department of Physics, University of Cambridge, with research interests including bacterial gene expression regulation, cilia synchronization in biological tissues, and control of self-assembly based on lipid systems; and Lorenzo Di Michele, affiliated with the Department of Chemistry, Faculty of Natural Sciences, Imperial College London, whose research involves DNA nanotechnology, self-assembly, and its applications in super-resolution optical microscopy.
Abstract
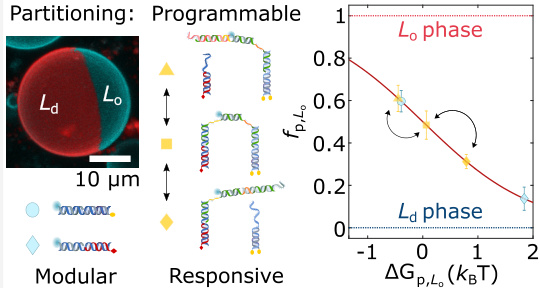
- A modular approach is introduced for programming the partitioning of amphiphilic DNA nanostructures within coexisting lipid domains.
- The lateral distribution of the devices is tuned by rational combination of hydrophobic anchors and alteration of the size and topology of the nanostructures, leveraging their tendency to enrich in different phases.
- The functionality of this strategy is demonstrated using a bio-inspired DNA architecture that dynamically undergoes ligand-induced restructuring to mediate cargo transport between domains through lateral redistribution.
- This lays the groundwork for the next generation of biomimetic platforms for sensing, transduction, and communication in synthetic cell systems.
Introduction
- Cell membranes regulate the distribution of biomachines within phaseseparated lipid domains to facilitate key processes such as signal transduction and transport.
- Underlying synthetic biology aims to replicate these life functions within artificial cell systems.
- A modular method for programming the partitioning of amphiphilic DNA nanostructures within coexisting lipid domains is introduced.
- The lateral distribution of the devices is regulated by rational combination of hydrophobic anchors and alterations in the size and topology of the nanostructures.
- The functionality of this strategy is demonstrated using a bio-inspired DNA architecture that undergoes dynamic ligand-induced restructuring through lateral redistribution to mediate cargo transport between domains.
- This paves the way for the next generation of biomimetic platforms for sensing, transduction, and communication in synthetic cell systems.
Materials and Methods
- Synthetic DNA oligonucleotides are used to construct nanostructures, some of which are covalently linked to hydrophobic groups to mediate membrane anchoring.
- Fluorescent tags (Alexa488, unless otherwise specified) are employed to monitor the distribution of nanostructures via confocal microscopy.
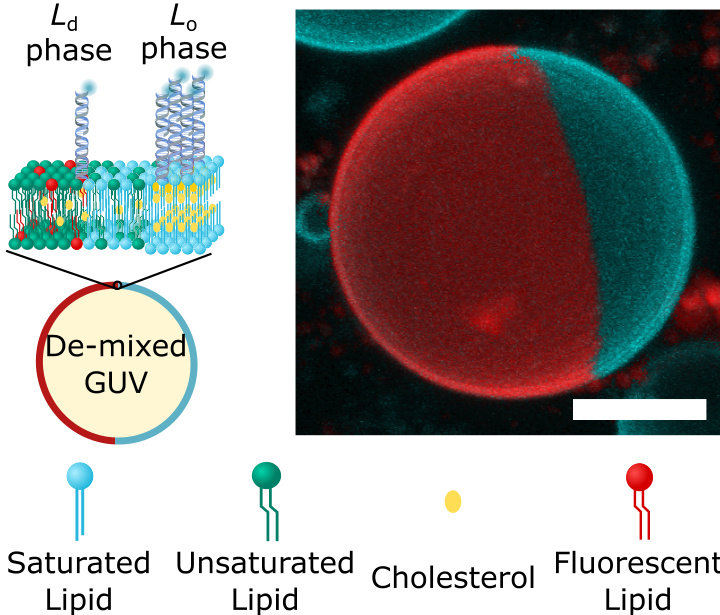
- Experiments are conducted using phase-separated giant unilamellar vesicles (GUVs) prepared from a ternary lipid mixture (DOPC/DPPC/sterol) to display Ld-Lo coexistence.
- GUVs are doped with TexasRed-DHPE to visualize lipid domains through confocal microscopy.
- A custom image processing workflow is used to analyze equatorial confocal micrographs of GUVs to quantify the lateral distribution of nanostructures.
- Förster Resonance Energy Transfer (FRET) and fluorescence signal crosstalk experiments are used to verify the proportional relationship between fluorescence intensity and nanostructure concentration.
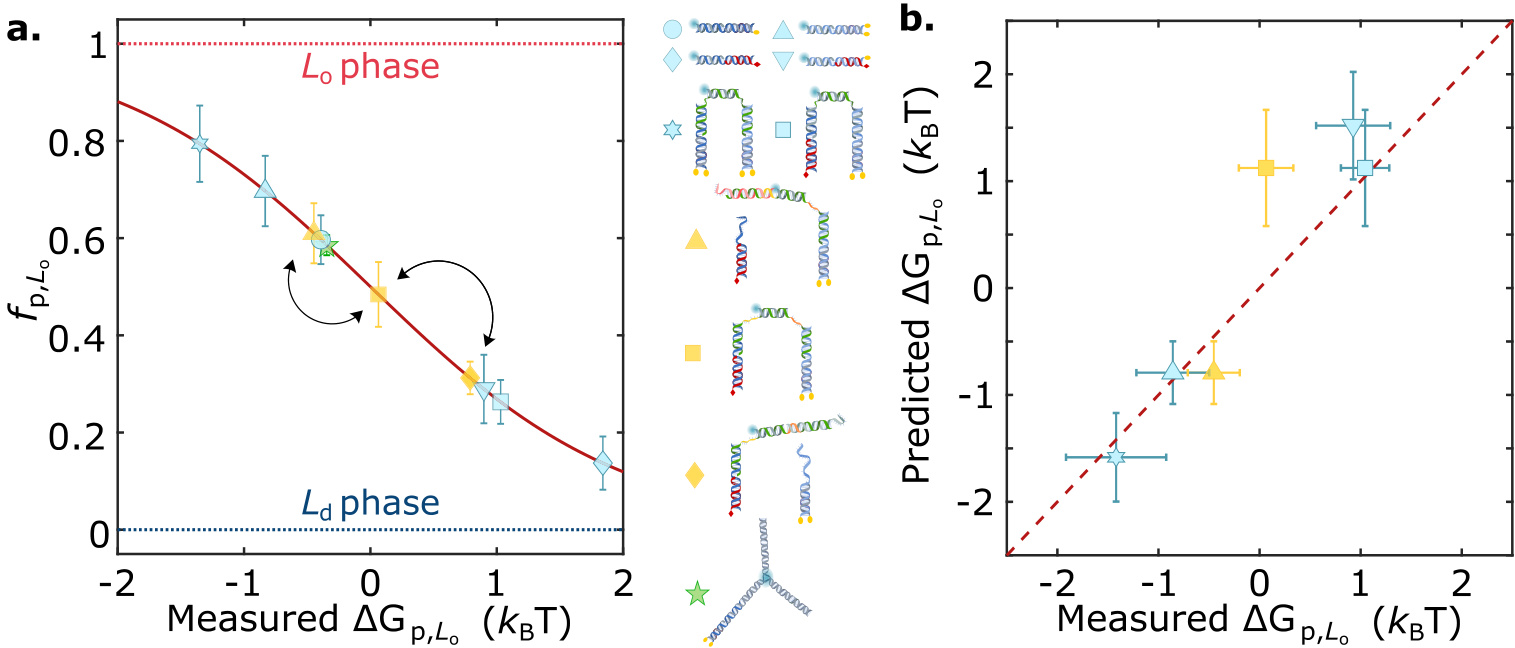
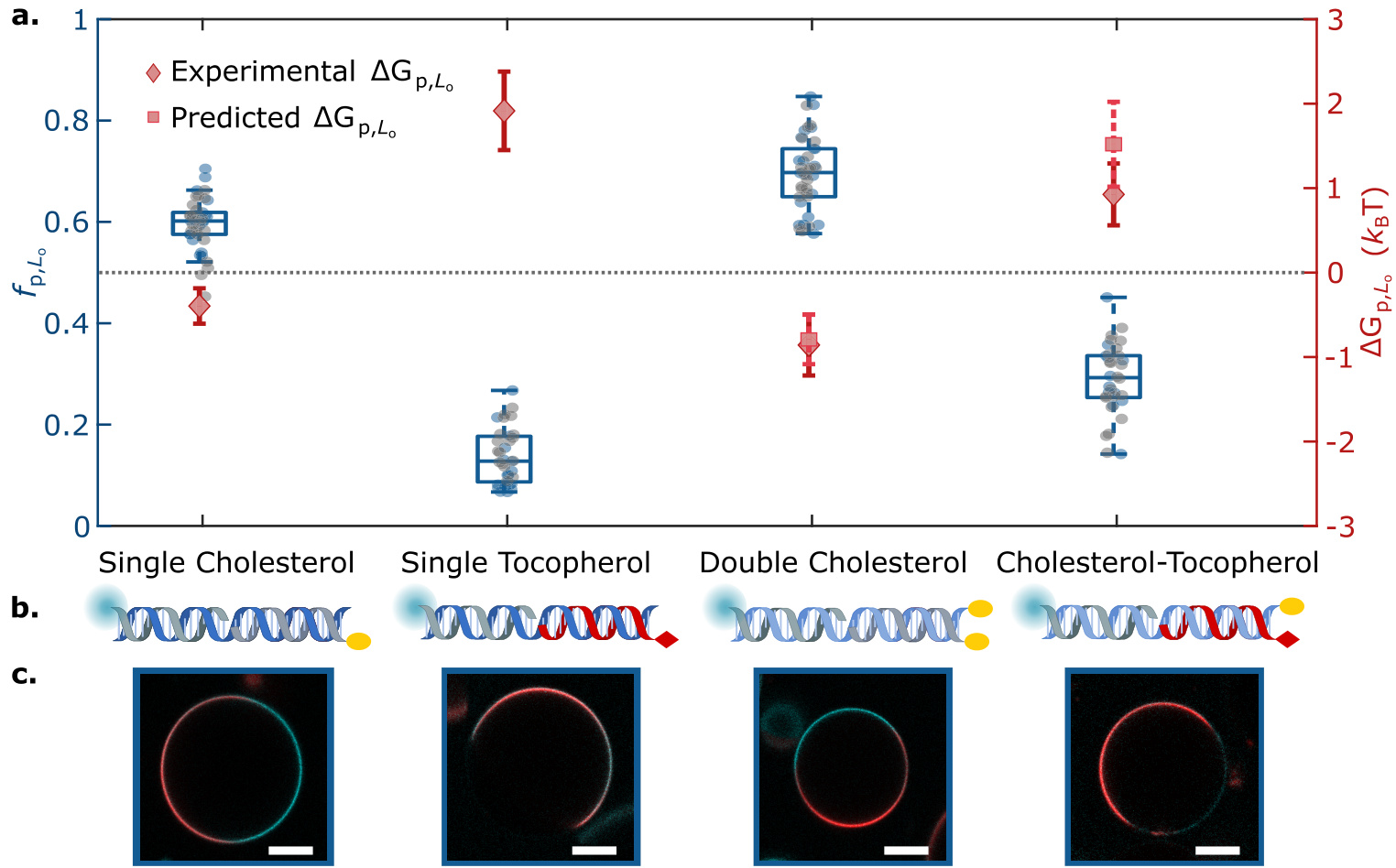
- Partition free energy of nanostructures is calculated using a formula.
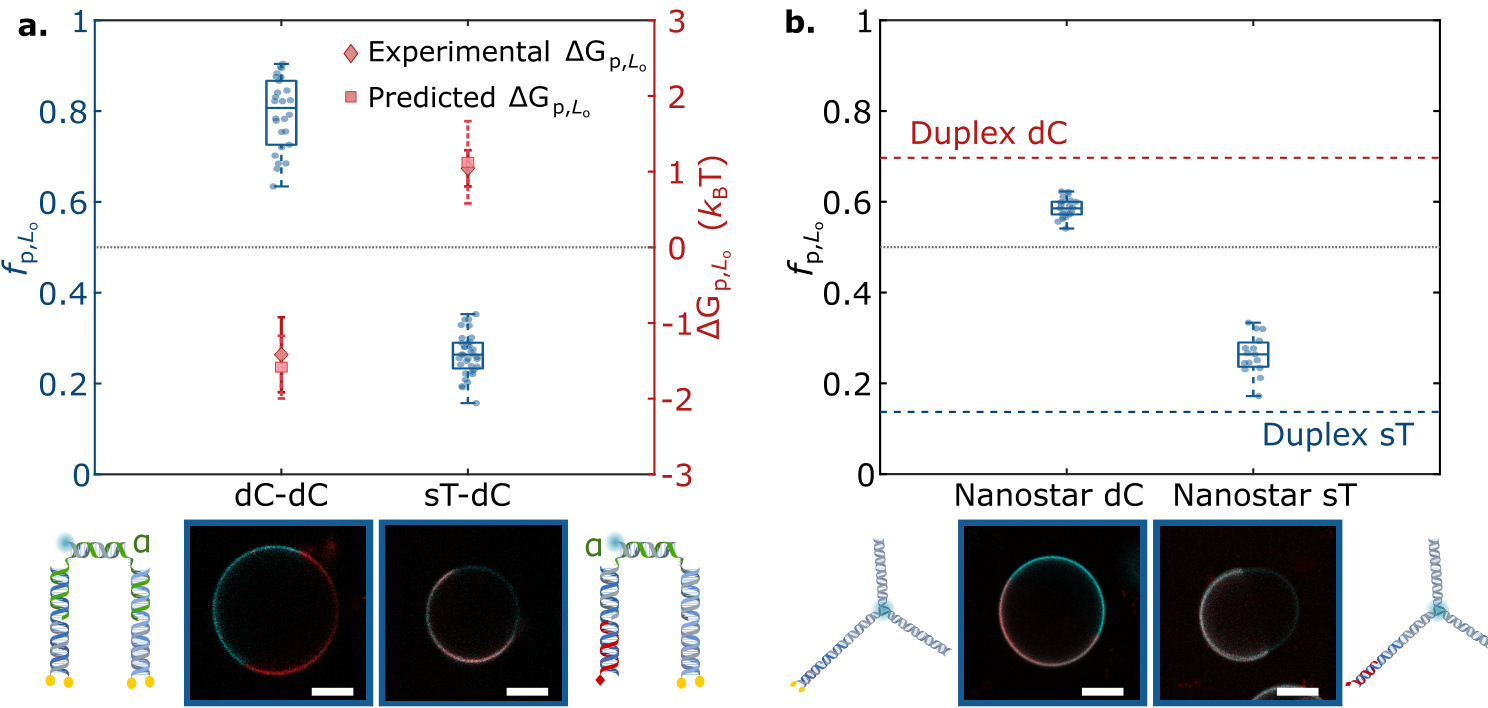
- DNA double strands with different hydrophobic anchors are used in experiments to study their partitioning behavior.
- DNA double strands with multiple hydrophobic anchors are used in experiments to study their partitioning behavior.
- DNA nanostructures with different topologies are used in experiments to study their partitioning behavior.
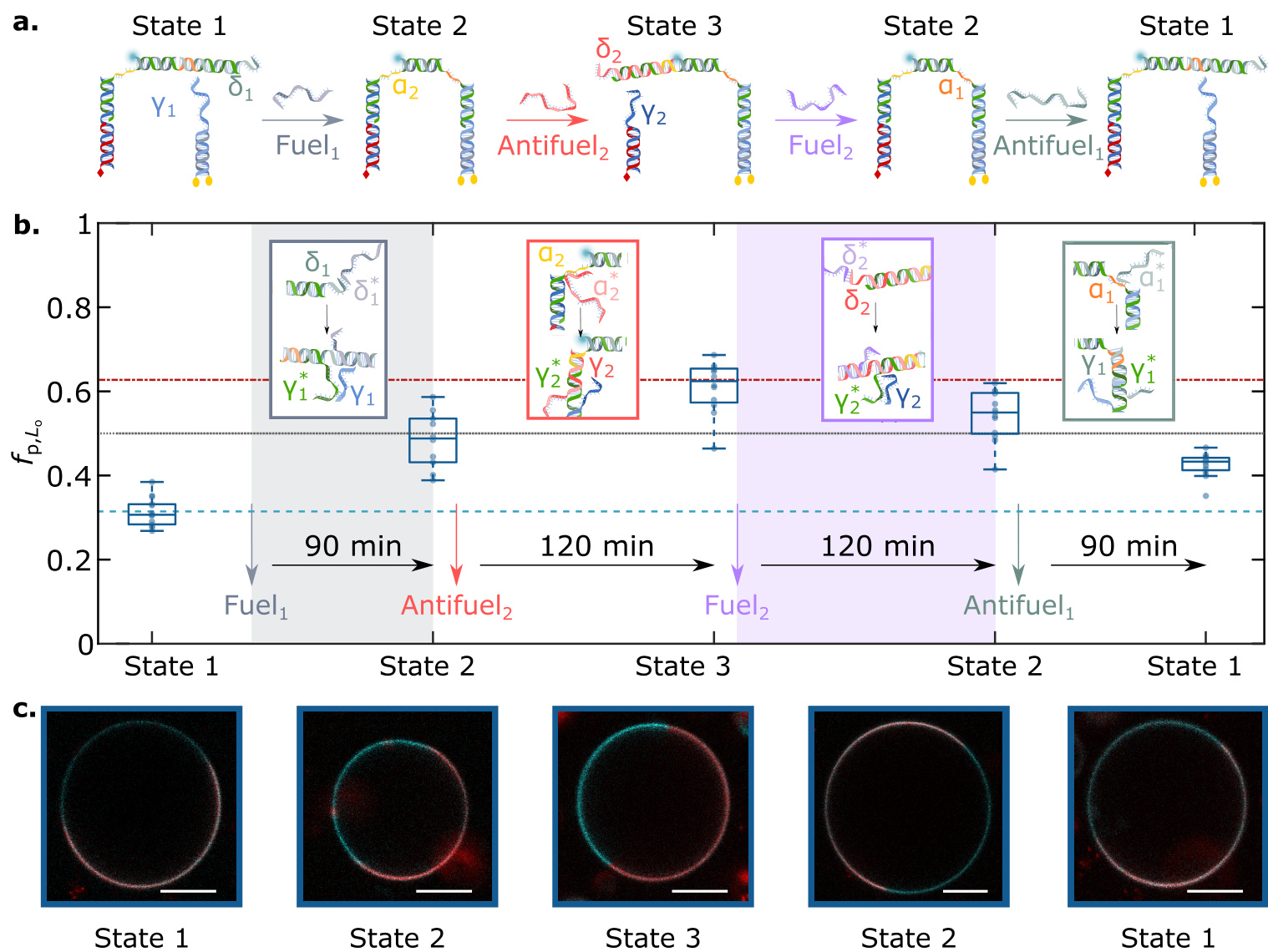
- DNA nanostructures with dynamic restructuring capabilities are used in experiments to study their cargo transport under molecular stimulation.
Results and Discussion
- The type and number of hydrophobic anchors affect the partitioning behavior of nanostructures.
- The partitioning behavior of nanostructures can be programmed by combining different hydrophobic anchors.
- The size and topology of nanostructures influence their partitioning behavior.
- The partitioning state of nanostructures can be dynamically controlled by altering the combination of anchors.
- Molecular stimulation can trigger dynamic restructuring of nanostructures, enabling cargo transport between lipid domains.
Conclusion
- A modular approach for programming the partitioning of amphiphilic DNA nanostructures within coexisting lipid domains is introduced.
- This method can be used to construct DNA nanostructures with dynamic restructuring capabilities to enable cargo transport between lipid domains.
- This platform paves the way for the next generation of biomimetic platforms for sensing, transduction, and communication in synthetic cell systems.
Q: What specific research methods were used in the paper?
- DNA Nanotechnology: Constructed DNA nanostructures with different hydrophobic anchor groups and topologies.
- Liposome Experiments: Utilized phase-separated giant unilamellar vesicles (GUVs) to simulate cell membrane environments and to study the distribution of DNA nanostructures within different lipid phases.
- Fluorescence Microscopy: Observed and quantified the distribution of DNA nanostructures in different lipid phases using confocal microscopy with fluorescence-labeled DNA nanostructures and liposomes.
- Image Analysis: Developed an image processing workflow to analyze confocal microscopy images and calculate the fluorescence intensity ratio (fp,Lo) of DNA nanostructures in different lipid phases.
- Free Energy Calculation: Calculated the free energy change (ΔGp,Lo) for the transfer of DNA nanostructures between different lipid phases based on fp,Lo values.
- Modular Design: Constructed DNA nanostructures with different phase distribution properties by combining different hydrophobic anchor groups and DNA nanostructures.
- Dynamic Reconfiguration: Demonstrated the dynamic reconfiguration capability of DNA nanostructures, allowing for the redistribution of fluorescent cargo on the surface of liposomes through molecular signaling.
Q: What are the main research findings and achievements?
- Modular Design Platform: Successfully constructed a modular platform that can precisely program the distribution of DNA nanostructures in phase-separated liposomes.
- Influence of Anchor Groups: Found that different hydrophobic anchor groups (such as cholesterol and tocopherol) significantly affect the phase distribution of DNA nanostructures.
- Free Energy Prediction: Developed a simple free energy prediction formula that can quantitatively predict the phase distribution of multi-anchored DNA nanostructures.
- Dynamic Reconfiguration: Demonstrated that DNA nanostructures can undergo dynamic reconfiguration triggered by molecular signals, thus enabling the redistribution of fluorescent cargo on the surface of liposomes.
Q: What are the current limitations of this research?
- Non-Additive Effects: In some cases, the phase distribution behavior of DNA nanostructures exhibits non-additive effects, which may be due to synergistic interactions between anchor groups or differences in lipid microenvironments.
- Exclusion Volume Effect: For larger DNA nanostructures, the exclusion volume effect may inhibit their accumulation in specific lipid phases, thereby reducing their phase distribution propensity.
- Saturation Effect: At high DNA/lipid ratios, liposomes may exhibit a saturation effect, preventing further accumulation of DNA nanostructures in the lipid phase.
- Dynamic Reconfiguration Efficiency: The efficiency of dynamic reconfiguration of DNA nanostructures may be influenced by various factors, such as the diffusion rate of fuel/antifuel strands and the rate of toehold-mediated strand displacement reactions.

被引用1372 | 浏览
被引用151 | 浏览
被引用981 | 浏览
被引用243 | 浏览
被引用332 | 浏览
被引用344 | 浏览
被引用747 | 浏览
被引用273 | 浏览
被引用69 | 浏览
被引用28 | 浏览
被引用110 | 浏览
被引用49 | 浏览